R
R has many tools for Bayesian analysis, and possessed these before Stan came around. Among the more prominent were those that allowed the use of BUGS (e.g. r2OpenBugs), one of its dialects JAGS (rjags), and packages like coda and MCMCpack that allowed for customized approaches, further extensions or easier implementation. Other packages might regard a specific type or family of models (e.g. bayesm), but otherwise be mostly R-like in specifying the model (e.g. MCMCglmm for mixed models).
Now it is as easy to conduct standard and more complex models using Stan while staying within the usual framework of R-style modeling. You don’t even have to write Stan code! I’ll later note a couple relevant packages that enable this.
rstan
The rstan package is the workhorse, and the other packages mentioned in following rely on it or assume similarities to it. In general though, rstan is what you will use when you write Stan code directly. The following demonstrates how.
Data list
First you’ll need to create a list of objects we’ll call the data list. It is a list of named objects that Stan will look to match to the things you noted in the data{} section of your Stan code. In our example, our data statement has four components- N K X and y. As such, we might create the following data list.
dataList = list(X=mymatrix, y=mytarget, N=1000, K=ncol(mymatrix))You could add fixed parameters and similar if your Stan code relies on them somewhere, but at this point you’re ready to proceed. Here is a model using RStan.
library(rstan)
modResults = stan(mystancode, data=dataList, iter=2000, warmup=1000)The Stan code is specified as noted previously, and can be a string in the R environment, or a separate text file7 I would maybe suggest using strings with simple models as you initially learn Stan/RStan, but a separate file is preferred. RStudio has syntax highlighting and other benefits for *.stan files..
Debug model
The debug model is just like any other except you’ll only want a couple iterations and for one chain.
model_debug = stan(mystancode, data=dataList, iter=10, chains=1)This will allow you to make sure the Stan code compiles first and foremost, and secondly, that there aren’t errors in the program that keep parameters from being estimated (thus resulting in no posterior samples). For a compile check, you hardly need any iterations. However, if you set the iterations a little higher, you may also discover potential difficulties in the estimation process that might suggest code issues remain.
Full model
If all is well with the previous, you can now proceed with the main model. Setting the argument fit = debugModel will save you the time spent compiling. It is a notable time saver to run the chains in parallel by setting cores = ncore, where ncore is some value representing the number of cores on your machine you want to use.
mystanmodel = stan(mystancode, data=dataList, fit = model_debug,
iter=2000, warmup=1000, thin=1, chains=4, cores=4)Once you are satisfied that the model runs well, you really only need one chain if you rerun it in the future.
Model summary
The typical model summary provides parameter estimates, standard errors, interval estimates and two diagnostics- effective sample size, the \(\hat{R}\) diagnostic.
print(mystanmodel, pars=c('Intercept', 'beta_1', 'sigma', 'Rsq'), probs = c(.05,.95), digits=3)Inference for Stan model: 34b5bc624f3e17e6f88e764a5bc0c898.
4 chains, each with iter=2000; warmup=1000; thin=1;
post-warmup draws per chain=1000, total post-warmup draws=4000.
mean se_mean sd 5% 95% n_eff Rhat
Intercept 1.267 0.002 0.085 1.126 1.403 1396 1.004
beta_1 0.081 0.004 0.148 -0.157 0.331 1606 1.003
sigma 0.969 0.001 0.030 0.922 1.019 2313 0.999
Rsq 0.646 0.000 0.001 0.644 0.648 1816 1.002
Samples were drawn using NUTS(diag_e) at Wed Nov 09 15:32:08 2016.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).Diagnostics and beyond
Typical Bayesian diagnostic tools like trace plots, density plots etc. are available. Part of the printed output contains the two just mentioned. In addition rstan comes with model comparison functions like WAIC and loo. The best part is the launch_shiny function, which actually makes this part of the analysis a lot more fun. Below is a graphical depiction of what would open in your browser when you use the function.
library(shinystan)
launch_shiny(mystanmodel)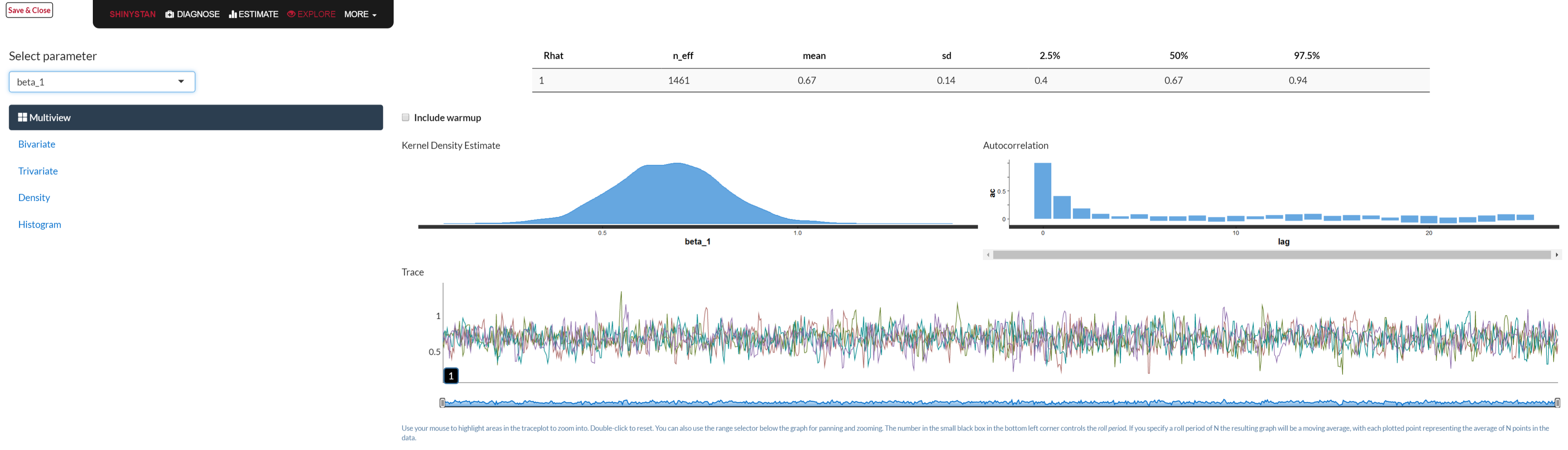
rstanarm
The rstanarm is a package from the Stan developers that allows you to specify models in the standard R formatThe ‘arm’ in rstanarm is for ‘applied regression and multilevel modeling’, which is NOT the title of Gelman’s book no matter what he says.. While this is very limiting, it definitely covers a lot of the usual statistical ground. As such, it enables you to be a Bayesian for any of the very common glm settings, including mixed and additive models.
Key modeling functions include:
- stan_lm: as with lm
- stan_glm: as with glm
- stan_glmer: generalized linear mixed models
- stan_gamm4: generalized additive mixed models
- stan_polr: ordinal regression models
Other functions allow the ability to change priors, enable posterior predictive checking etc. The following shows example code.
library(rstanarm)
rstanarm_results = stan_glm(y ~ x, data=mydataframe, iter=2000, warmup=1000, cores=4)
summary(rstanarm_results, probs=c(.025, .975), digits=3)stan_glm(formula = y ~ x, data = mydataframe, iter = 2000, warmup = 1000)
Family: gaussian (identity)
Algorithm: sampling
Posterior sample size: 4000
Observations: 500
Estimates:
mean sd 2.5% 97.5%
(Intercept) 1.272 0.086 1.101 1.441
x 0.071 0.148 -0.216 0.364
sigma 0.969 0.030 0.912 1.030
mean_PPD 1.309 0.060 1.189 1.426
log-posterior -700.510 1.131 -703.489 -699.217
Diagnostics:
mcse Rhat n_eff
(Intercept) 0.001 1.000 3298
x 0.003 0.999 3364
sigma 0.001 0.999 3428
mean_PPD 0.001 1.000 3553
log-posterior 0.022 1.001 2598
For each parameter, mcse is Monte Carlo standard error, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence Rhat=1).The resulting model object can essentially be used just like the lm/glm functions in base R. There are summary, predict, fitted, coef etc. functions available to use just like with standard model objects.
brms
I have watched with much enjoyment the development of the brms package from nearly its inception. Due to the continued development of rstanarm, it’s role is becoming more niche perhaps, but I still believe it to be both useful and powerful. It allows for many types of models, custom Stan functions, and many distributions (including truncated versions, ordinal variables, zero-inflated, etc.). The main developer is ridiculously responsive to requests, so extensions are regularly implemented. In short, for standard models you can use rstanarm, while for variations of those, more flexible manipulation of priors, or more complex models, you can use brms.
The following shows an example of the additional capabilities provided by the brm function, which unlike rstanarm, is the only function you need for modeling with this package. The following demonstrates use of a truncated distribution, an additive model with random effect, use of a different family function, specification of prior distribution for the fixed effect coefficients, specification of correlated residual structure, optional estimation algorithm, and use of custom Stan functions.
library(brms)
modResults = brm(y | trunc(lb = 0, ub = 100) ~ s(x) + (1|id), family=student, data=dataList,
prior = set_prior('horseshoe(3)', class='b'),
autocor = cor_ar(~patient, p = 1),
algorithm = 'meanfield',
stan_funs = stdized,
iter=2000, warmup=1000)The brms package also has a lot of the same functionality for post model inspection.
rethinking
The rethinking package accompanies the text, Statistical Rethinking by Richard McElreath. This is a great resource for learning Bayesian data analysis while using Stan under the hood. You can do more with the other packages mentioned, but if you can also run your model here, you might get even more to play with. Like rstanarm and brms, you might be able to use it to produce starter Stan code as well, that you can then manipulate and use via rstan. Again, this is a very useful tool to learn Bayesian analysis in general, especially if you have the text.
Summary
To recap, we can summarize the roles of these packages as follows (ordered from easiest to more flexible):
rethinking: Good resource to introduce yourself to Bayesian analysis.
rstanarm: All you need to start on your path to using Bayesian techniques. Keeps you in more familiar modeling territory so you can focus on learning the new stuff. Supports up through mixed models, GAMs, and ordinal models.
brms: Take your model notably further and still not have to write raw stan code. Supports a very wide range of models and still without raw Stan code.
rstan: Here you write your Stan code regarding whatever model your heart desires, then let rstan do the rest.
raw R or other: Some still insist on writing their own sampler. While great for learning purposes, this is mostly a good way to produce buggier code, have less model exploration capability, all while being a lot less efficient (and very slow if in raw R). Maybe you’ll end up here, but you should exhaust your other possibilities first.